Publications
We conduct research to bridge the gap between life sciences and computer science, employing advanced sequencing technologies to analyze (meta-)genomic and transcriptomic datasets related to infectious diseases, immunology, and medical sciences. With this interdisciplinary approach, we aim to improve our understanding of pathogen-host interactions and support genomic surveillance and containment. We perform research in the following areas:
- Microbial bioinformatics
- Genomic surveillance
- Pathogen detection and characterization
- Nanopore real-time sequencing
- Transcriptomics and gene expression
- Metagenomics and metatranscriptomics
- Reproducible genomics (container, WMS)
Highlighted
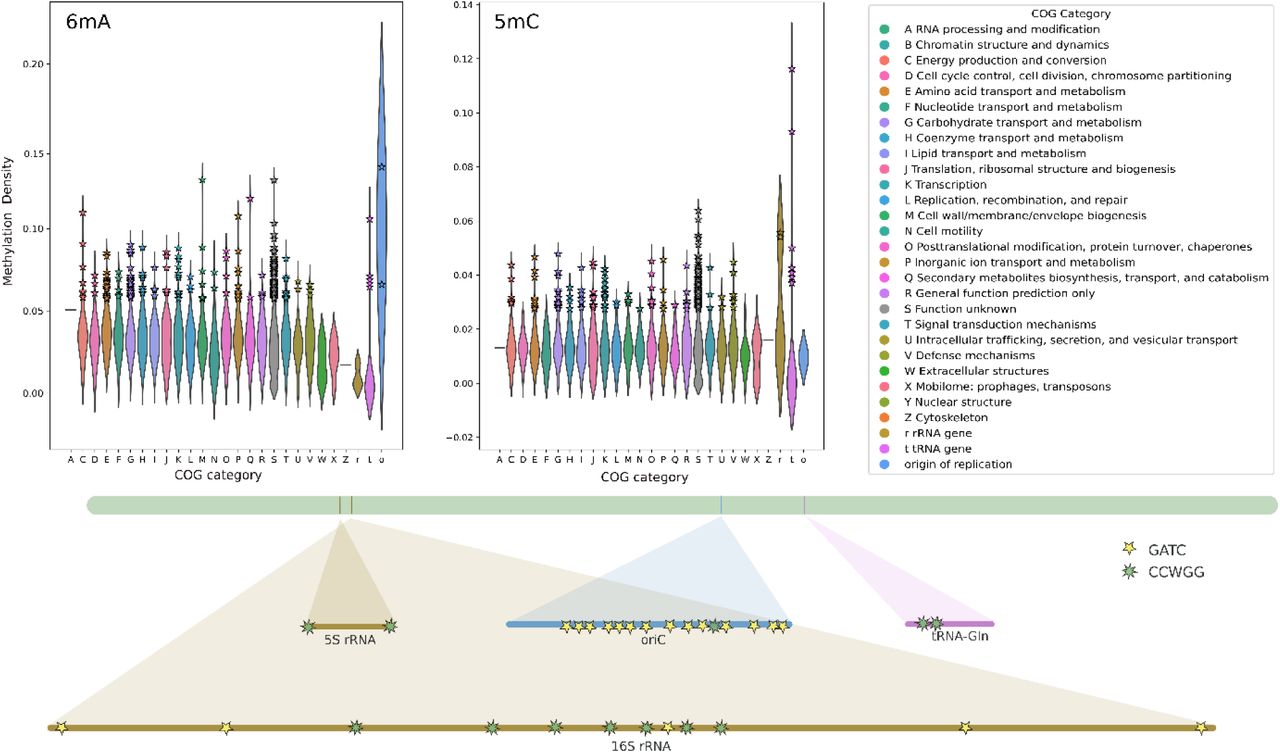
Decoding bacterial methylomes in four public health-relevant microbial species: Nanopore sequencing enables reproducible analysis of DNA modifications
openRxiv
·
02 Feb 2025
·
doi:10.1101/2025.01.29.635492
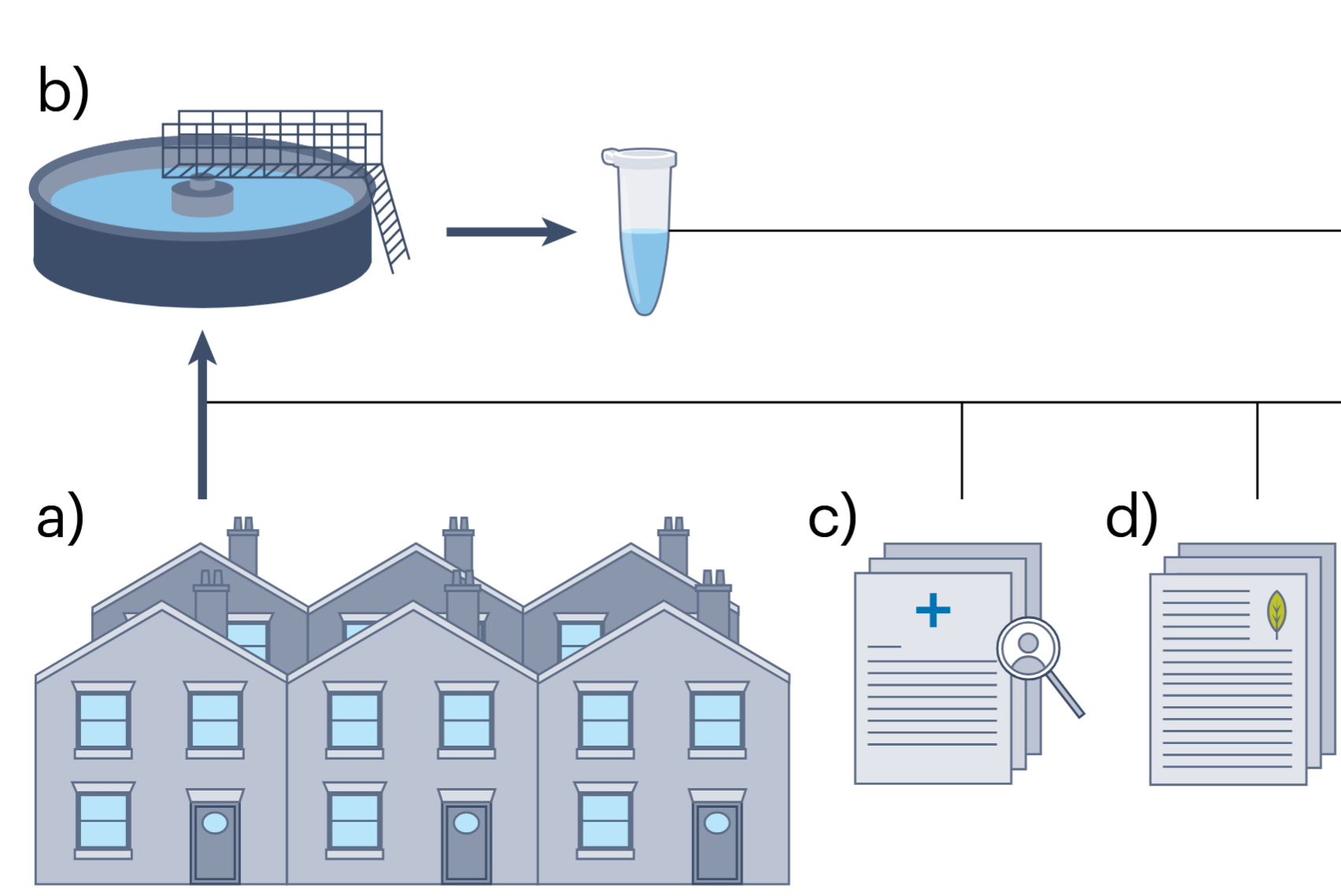
Augmentation of wastewater-based epidemiology with machine learning to support global health surveillance
Nature Water
·
02 Jun 2025
·
doi:10.1038/s44221-025-00444-5
All
2026
Addressing pandemic-wide systematic errors in the SARS-CoV-2 phylogeny
Nature Methods
·
09 Feb 2026
·
doi:10.1038/s41592-025-02947-1
2025
A whole-genome sequencing dataset of nanopore raw signals for bacterial genotyping and methylation analysis
Scientific Data
·
02 Dec 2025
·
doi:10.1038/s41597-025-06319-4
A public health perspective of SARS-CoV-2 evolution and surveillance strategies in Germany from 2020 to 2023
Communications Medicine
·
11 Nov 2025
·
doi:10.1038/s43856-025-01093-1
Genomic Donor-Recipient Mismatches in Kidney Transplantation: A Focus on the Techniques and Approaches
Current Transplantation Reports
·
30 Aug 2025
·
doi:10.1007/s40472-025-00492-6
Accurately assembling nanopore sequencing data of highly pathogenic bacteria
BMC Genomics
·
28 Aug 2025
·
doi:10.1186/s12864-025-11793-6
Large-scale Evaluation of Prokaryotic Annotation Tools Across Thousands of Species
Springer Science and Business Media LLC
·
13 Aug 2025
·
doi:10.21203/rs.3.rs-7317585/v1
Real-time, multi-pathogen wastewater genomic surveillance with Freyja 2
Cold Spring Harbor Laboratory
·
27 Jul 2025
·
doi:10.1101/2025.07.26.25332245
Next-Generation Sequencing Analysis in Greek Patients with Predominantly Antibody Deficiencies
Immuno
·
16 Jul 2025
·
doi:10.3390/immuno5030027
In silico genomic surveillance by CoVerage predicts and characterizes SARS-CoV-2 variants of interest
Nature Communications
·
08 Jul 2025
·
doi:10.1038/s41467-025-60231-4
Targeted decontamination of sequencing data with CLEAN
NAR Genomics and Bioinformatics
·
04 Jul 2025
·
doi:10.1093/nargab/lqaf105

Augmentation of wastewater-based epidemiology with machine learning to support global health surveillance
Nature Water
·
02 Jun 2025
·
doi:10.1038/s44221-025-00444-5
varVAMP: degenerate primer design for tiled full genome sequencing and qPCR
Nature Communications
·
31 May 2025
·
doi:10.1038/s41467-025-60175-9
Leveraging wastewater sequencing to strengthen global public health surveillance
BMC Global and Public Health
·
21 Mar 2025
·
doi:10.1186/s44263-025-00138-w

Decoding bacterial methylomes in four public health-relevant microbial species: Nanopore sequencing enables reproducible analysis of DNA modifications
openRxiv
·
02 Feb 2025
·
doi:10.1101/2025.01.29.635492
SARS-CoV-2 evolution on a dynamic immune landscape
Nature
·
29 Jan 2025
·
doi:10.1038/s41586-024-08477-8
Exploring the genomic landscape of Chlamydiifrater species: Novel features include multiple truncated major outer membrane proteins (MOMP), unique genes, and chlamydial plasticity zone orthologs
Microbiology Society
·
06 Jan 2025
·
doi:10.1099/acmi.0.000936.v2
VirusWarn: A mutation-based early warning system to prioritize concerning SARS-CoV-2 and influenza virus variants from sequencing data
Computational and Structural Biotechnology Journal
·
01 Jan 2025
·
doi:10.1016/j.csbj.2025.03.010
Exploring the genomic landscape of Chlamydiifrater species: novel features include multiple truncated major outer membrane proteins, unique genes and chlamydial plasticity zone orthologs
Access Microbiology
·
01 Jan 2025
·
doi:10.1099/acmi.0.000936.v3
2024
Systematic Screening of Autosomal Dominant Tubulointerstitial Kidney Disease–MUC1 27dupC Pathogenic Variant through Exome Sequencing
Journal of the American Society of Nephrology
·
26 Sep 2024
·
doi:10.1681/ASN.0000000503
CIEVaD: A Lightweight Workflow Collection for the Rapid and On-Demand Deployment of End-to-End Testing for Genomic Variant Detection
Viruses
·
11 Sep 2024
·
doi:10.3390/v16091444
Endogenous Bornavirus-like Elements in Bats: Evolutionary Insights from the Conserved Riboviral L-Gene in Microbats and Its Antisense Transcription in Myotis daubentonii
Viruses
·
27 Jul 2024
·
doi:10.3390/v16081210
RIBAP: a comprehensive bacterial core genome annotation pipeline for pangenome calculation beyond the species level
Genome Biology
·
01 Jul 2024
·
doi:10.1186/s13059-024-03312-9
Lessons learned: overcoming common challenges in reconstructing the SARS-CoV-2 genome from short-read sequencing data via CoVpipe2
F1000Research
·
16 Apr 2024
·
doi:10.12688/f1000research.136683.2
POCP-nf: an automatic Nextflow pipeline for calculating the percentage of conserved proteins in bacterial taxonomy
Bioinformatics
·
29 Mar 2024
·
doi:10.1093/bioinformatics/btae175
ICTV Virus Taxonomy Profile: Kolmioviridae 2024
Journal of General Virology
·
29 Feb 2024
·
doi:10.1099/jgv.0.001963
Reannotation of the zebra finch genome reveals undiscovered transcriptional complexity
openRxiv
·
09 Jan 2024
·
doi:10.1101/2024.01.09.574630
Impact of reference design on estimating SARS-CoV-2 lineage abundances from wastewater sequencing data
GigaScience
·
01 Jan 2024
·
doi:10.1093/gigascience/giae051
2023
Unlocking the Full Potential of Nanopore Sequencing: Tips, Tricks, and Advanced Data Analysis Techniques
openRxiv
·
07 Dec 2023
·
doi:10.1101/2023.12.06.570356
Dicer‐dependent heterochromatic small RNAs in the model diatom species Phaeodactylum tricornutum
New Phytologist
·
03 Dec 2023
·
doi:10.1111/nph.19429
decOM: similarity-based microbial source tracking of ancient oral samples using k-mer-based methods
Microbiome
·
06 Nov 2023
·
doi:10.1186/s40168-023-01670-3
aKmerBroom: Ancient oral DNA decontamination using Bloom filters on k-mer sets
iScience
·
01 Nov 2023
·
doi:10.1016/j.isci.2023.108057
ICTV Virus Taxonomy Profile: Xinmoviridae 2023
Journal of General Virology
·
26 Oct 2023
·
doi:10.1099/jgv.0.001906
VIRify: An integrated detection, annotation and taxonomic classification pipeline using virus-specific protein profile hidden Markov models
PLOS Computational Biology
·
28 Aug 2023
·
doi:10.1371/journal.pcbi.1011422
Neighbourhood watch: genomic epidemiology of SARS-CoV-2 variants circulating in a German federal state, Mecklenburg-Western Pomerania, in 2020–2022
Emerging Microbes & Infections
·
22 Aug 2023
·
doi:10.1080/22221751.2023.2245916
Annual (2023) taxonomic update of RNA-directed RNA polymerase-encoding negative-sense RNA viruses (realm
The Journal of general virology
·
01 Aug 2023
·
pmid:37622664
ICTV Virus Taxonomy Profile: Lispiviridae 2023
Journal of General Virology
·
11 Jul 2023
·
doi:10.1099/jgv.0.001869
Building alternative splicing and evolution-aware sequence-structure maps for protein repeats
openRxiv
·
30 Apr 2023
·
doi:10.1101/2023.04.29.538821
De novo genome assembly resolving repetitive structures enables genomic analysis of 35 European Mycoplasma bovis strains
openRxiv
·
13 Apr 2023
·
doi:10.1101/2023.04.13.536562
Sequencing accuracy and systematic errors of nanopore direct RNA sequencing
openRxiv
·
30 Mar 2023
·
doi:10.1101/2023.03.29.534691
Nanopore-based enrichment of antimicrobial resistance genes – a case-based study
Gigabyte
·
25 Jan 2023
·
doi:10.46471/gigabyte.75
Managing and monitoring a pandemic: showcasing a practical approach for the genomic surveillance of SARS-CoV-2
Database
·
01 Jan 2023
·
doi:10.1093/database/baad071
2022
Women in the European Virus Bioinformatics Center
Viruses
·
12 Jul 2022
·
doi:10.3390/v14071522
CovRadar: continuously tracking and filtering SARS-CoV-2 mutations for genomic surveillance
Bioinformatics
·
07 Jul 2022
·
doi:10.1093/bioinformatics/btac411
Advancing Precision Vaccinology by Molecular and Genomic Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 in Germany, 2021
Clinical Infectious Diseases
·
25 Jun 2022
·
doi:10.1093/cid/ciac399
Synopse virologischer Analysen im Nationalen Referenzzentrum für Influenzaviren während der COVID-19-Pandemie
Robert Koch-Institut
·
02 Jun 2022
·
doi:10.25646/10118
History and classification of Aigai virus (formerly Crimean–Congo haemorrhagic fever virus genotype VI)
Journal of General Virology
·
12 Apr 2022
·
doi:10.1099/jgv.0.001734
Rapid characterization of a Delta-Omicron SARS-CoV-2 recombinant detected in Europe
Research Square
·
04 Apr 2022
·
doi:10.21203/rs.3.rs-1502293/v1
Jingchuvirales: a New Taxonomical Framework for a Rapidly Expanding Order of Unusual Monjiviricete Viruses Broadly Distributed among Arthropod Subphyla
Applied and Environmental Microbiology
·
22 Mar 2022
·
doi:10.1128/aem.01954-21
Near Chromosome-Level Genome Assembly and Annotation of Rhodotorula babjevae Strains Reveals High Intraspecific Divergence
Journal of Fungi
·
22 Mar 2022
·
doi:10.3390/jof8040323
ASES: visualizing evolutionary conservation of alternative splicing in proteins
Bioinformatics
·
21 Feb 2022
·
doi:10.1093/bioinformatics/btac105
Investigation of a Limited but Explosive COVID-19 Outbreak in a German Secondary School
Viruses
·
04 Jan 2022
·
doi:10.3390/v14010087
What the Phage: a scalable workflow for the identification and analysis of phage sequences
GigaScience
·
01 Jan 2022
·
doi:10.1093/gigascience/giac110
2021
Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions
Nature Microbiology
·
24 Dec 2021
·
doi:10.1038/s41564-021-01011-w
Context-aware genomic surveillance reveals hidden transmission of a carbapenemase-producing Klebsiella pneumoniae
Microbial Genomics
·
24 Dec 2021
·
doi:10.1099/mgen.0.000741
2021 Taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales.
Archives of virology
·
01 Dec 2021
·
pmid:34463877
ICTV Virus Taxonomy Profile: Nyamiviridae 2021
Journal of General Virology
·
05 Nov 2021
·
doi:10.1099/jgv.0.001681
Rise and Fall of SARS-CoV-2 Lineage A.27 in Germany
Viruses
·
29 Jul 2021
·
doi:10.3390/v13081491
The economical lifestyle of CPR bacteria in groundwater allows little preference for environmental drivers
openRxiv
·
29 Jul 2021
·
doi:10.1101/2021.07.28.454184
Evidence for the existence of a new genus Chlamydiifrater gen. nov. inside the family Chlamydiaceae with two new species isolated from flamingo (Phoenicopterus roseus): Chlamydiifrater phoenicopteri sp. nov. and Chlamydiifrater volucris sp. nov.
Systematic and Applied Microbiology
·
01 Jul 2021
·
doi:10.1016/j.syapm.2021.126200
Assessing conservation of alternative splicing with evolutionary splicing graphs
Genome Research
·
15 Jun 2021
·
doi:10.1101/gr.274696.120
A global metagenomic map of urban microbiomes and antimicrobial resistance.
Cell
·
26 May 2021
·
pmid:34043940
poreCov - an easy to use, fast, and robust workflow for SARS-CoV-2 genome reconstruction via nanopore sequencing
openRxiv
·
07 May 2021
·
doi:10.1101/2021.05.07.443089
Metagenomics workflow for hybrid assembly, differential coverage binning, metatranscriptomics and pathway analysis (MUFFIN)
PLOS Computational Biology
·
09 Feb 2021
·
doi:10.1371/journal.pcbi.1008716
RUNX3 Transcript Variants Have Distinct Roles in Ovarian Carcinoma and Differently Influence Platinum Sensitivity and Angiogenesis
Cancers
·
26 Jan 2021
·
doi:10.3390/cancers13030476
SLC35F2, a Transporter Sporadically Mutated in the Untranslated Region, Promotes Growth, Migration, and Invasion of Bladder Cancer Cells
Cells
·
06 Jan 2021
·
doi:10.3390/cells10010080
2020
RNAflow: An Effective and Simple RNA-Seq Differential Gene Expression Pipeline Using Nextflow
Genes
·
10 Dec 2020
·
doi:10.3390/genes11121487
The International Virus Bioinformatics Meeting 2020
Viruses
·
06 Dec 2020
·
doi:10.3390/v12121398
Assessing Conservation of Alternative Splicing with Evolutionary Splicing Graphs
openRxiv
·
15 Nov 2020
·
doi:10.1101/2020.11.14.382820
A decade of de novo transcriptome assembly: Are we there yet?
Molecular Ecology Resources
·
29 Oct 2020
·
doi:10.1111/1755-0998.13268
Targeted domain assembly for fast functional profiling of metagenomic datasets with S3A.
Bioinformatics (Oxford, England)
·
01 Jul 2020
·
pmid:32330240
Transcripts’ Evolutionary History and Structural Dynamics Give Mechanistic Insights into the Functional Diversity of the JNK Family
Journal of Molecular Biology
·
01 Mar 2020
·
doi:10.1016/j.jmb.2020.01.032